otivation for
calculations of this section was presented in the previous section
(
Calculation of
approximation spaces
).
We start from the pair
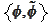 introduced in the section
(
Symmetric biorthogonal
wavelets
) and calculated in the section
(
Calculation of scaling
functions
). We intend to adapt the procedure of the section
(
Adapting GMRA to interval
[0,1]
) to have two properties:
introduced in the section
(
Symmetric biorthogonal
wavelets
) and calculated in the section
(
Calculation of scaling
functions
). We intend to adapt the procedure of the section
(
Adapting GMRA to interval
[0,1]
) to have two properties:
1. The procedure should be numerically stable.
2. The Gram
matrix
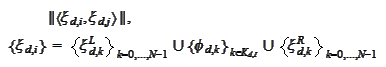 should have a low condition number.
should have a low condition number.
The following is the procedure that accomplishes these two goals.
The procedure is implemented in the script "bases\phi.py" and tested by the
script "_run_bases.py". Experimentation shows that for 6 steps of scaling
procedure we produce Gram matrix with condition number below 10 and precision
of scaling projection below 0.0001.
|